Genomics of Evolution and Development
Our research focusses on understanding the genomic basis of evolution and development in humans, as well as non-model organisms and traditional model species. We study how evolution has shaped our disease susceptibility using machine-learning approaches, and the robustness of gene-regulatory networks to mutational and environmental disruption. We use comparative genomics and epigenetic approaches to uncover how early embryogenic specification events govern adult morphology variation. Combining state-of-the-art computational, molecular, and imaging technologies we investigate the following topics:
- Comparative and developmental genomics. Comparative genomics is a powerful approach to unravel the underpinnings of species diversification. We investigate the evolutionary conservation of early embryonic development and neurophysiology and study the effects of gene duplication events on phenotypic robustness. To this end we combine high-throughput sequencing with genetic engineering and imaging in an array of systems ranging from annelids to echinoderms and zebrafish. Other areas of research focus on comparisons of ancient and present genomics to investigate extinction events and past epidemics in diverse species. We develop and apply machine-learning algorithms to infer human evolutionary admixture events and gene selection mechanisms.
- Epigenetics and phenotypic plasticity. How epigenetic mechanisms integrate environmental cues to govern phenotypic plasticity is a central question in gene regulation. We combine genomics approaches with non-traditional model systems such as ants, honeybee and crustaceans to uncover the mechanisms of polyphenism and adaptive changes. A further focus is on evolution of epigenetic regulators across eukaryotic lineages and their interaction with transposable elements.
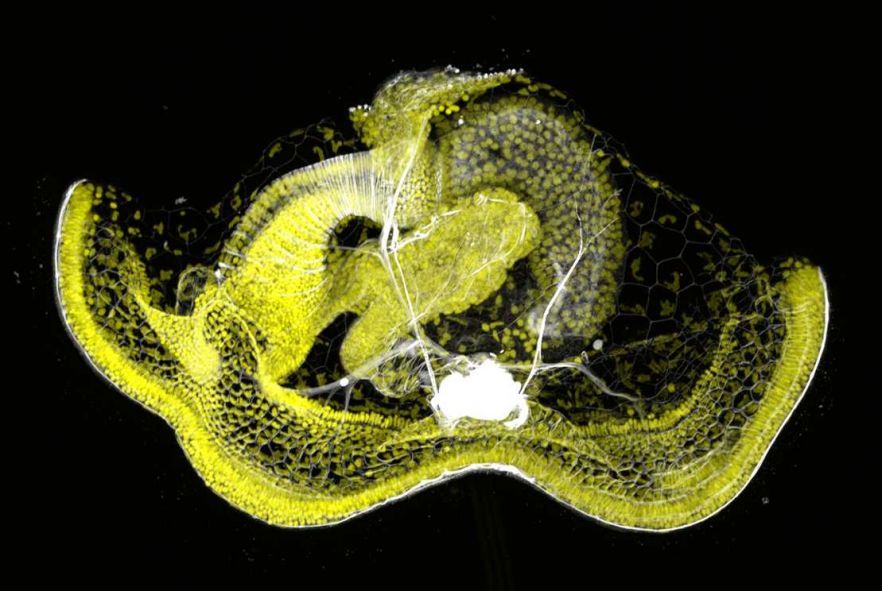 Mitraria larva of Owenia fusiformis (Annelida) stained for nuclei (yellow) and muscles (white). Credit: Allan Carrillo-Baltodano
Mitraria larva of Owenia fusiformis (Annelida) stained for nuclei (yellow) and muscles (white). Credit: Allan Carrillo-Baltodano
